|
Home |
|
Projects |
|
Landmarks |
|
Publications |
|
People |
|
Protocols |
|
Awards |
|
Research Supports |
|
Gallery |
|
Supplements |
|
Contact |


|
JAYARAM LAB |
|
Landmark discoveries/accomplishments Site-specific Recombination
1. 1990-1992: A single active site in the Flp recombianse is assembled from two monomers of the protein. Dr. Jing-Wen Chen and Mr. Jehee Lee, with help from Dr. Marie-Claude Serre make the ground breaking discovery that the active site of the Flp recombinase is assembled from two protein monomers. As a result the active site nucleophile Tyr-343 is delivered in trans to the scissile phopshate. Chen, J-W., Lee, J. and Jayaram, M. (1992). DNA cleavage in trans by the active site tyrosine during Flp recombination: switching protein partners before exchanging strands.
2. 1992-1994. The trans cleavage model is tested and verified. DNA cleavage at a scissile phosphate oriented by a Flp monomer can be effected by an exogenous nucleophile. Mr. Jehee Lee, Mr. Sang-Hwa Yang and Dr. Jing-Wen Chen use various catalytic mutants of Flp in pairwise combinations to rigorously establish the validity of the trans cleavage model. Dr. Lee demonstrates that an exogenous nucleophile such as tyramine or hydrogen peroxide can be delivered to a Flp mutant lacking Tyr-343 (the native cleavage nucleophile) to effect site-specific strand cutting. Chen, J-W., Yang, S-H.. and Jayaram, M. (1993). Tests for the fractional active-site model in Flp site-specific recombination. Assembly of a functional recombination complex in half-site and full-site strand transfer. J. Biol. Chem. 268: 14417-14425. Kimball, A. S., Lee, J., Jayaram, M. and Tullius, T. D. (1993). Sequence-specific cleavage of DNA via nucleophilic attack of hydrogen peroxide, assisted by Flp recombinase. BIochemistry 32: 469-4701.
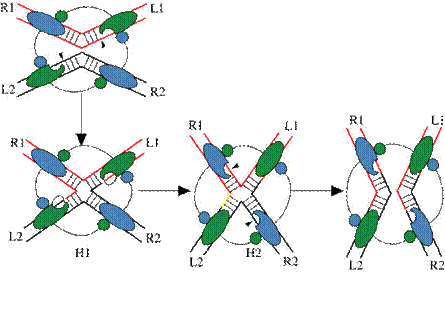
3. 1995-1997. The Holliday junction intermediate in Flp recombination is nearly square planar. Mr. Jehee Lee finds that the Flp recombinase, upon binding to synthetic Holliday junctions, imposes an antiparallel square planar geometry on them. Mr.Lee is assisted in this work by Dr. Yuri Voziyanov and Ms. Shailja Pathania. Lee, J., Voziyanov, Y., Pathania, S., and Jayaram, M. (1998). Structural alterations and conformational dynamics in Holliday junctions induced by binding of a site-specific recombinase. Mol. Cell 1: 483-493.
4. 1995-1997. Biochemical studies reveal the structural basis for the two-step strand exchange mechanism of recombination. Mr. Jehee Lee and Dr. Takashi Tonozuka design DNA substrates that are predisposed to bend one way or the other to show that the orientation of the bend determines which of the two possible active sites within a Flp dimer will be active. The bend induced 'half-of-the sites' activity explains the completion of recombination in two steps of single strand exchanges, and is supported by the crystal structures. Lee, J., Tonozuka, T. and Jayaram, M. (1997). Mechanism of active site exclusion in a site-specific recombinase: role of the DNA substrate in conferring half-of-the-sites activity. Genes and Dev. 11: 3061-3071. Lee, J. and Jayaram, M. (1997). A tetramer of Flp silences the trimers within it during resolution of a Holliday junction substrate. Genes and Dev. 11: 2438-2447.
5. 1997-2001. The Flp DNA recombinase contains cryptic RNA cleavage active sites. Dr. Chong-Jun Xu identifies two distinct types of RNA cleavage activities carried out by the Flp recombinase. The results suggest possible evolutionary routes for the assembly of the recombination active site from an elementary nuclease active site. Xu, C. J., Grainge, I., Lee, J., Harshey, R. M. and Jayaram, M. (1998). Unveiling two distinct ribonuclease activities and a topoisomerase activity in a site-specific DNA recombinase. Mol. Cell 1: 729-739. Sau, A. K., Tribble, G., Grainge, I., Frohlich, R. F., Knudsen, B. R., and Jayaram, M. (2001). Biochemical and kinetic analysis of the RNase activities of the Integrase/Tyrosine family recombinases. J. Biol. Chem. 276: 46612-46623
6. 1999-2000. Within the Flp recombination complex, the target DNA partners have an antiparallel orientation. Dr. Ian Grainge uses a topological assay to determine that the Flp recombinase acts on target sites arranged in an antiparallel fashion. No DNA crossing is introduced as a result of strand exchange. This result suggests the use of Flp (or the related recombinase Cre) for mapping the DNA path in any uncharacterized DNA-protein assembly. The method is termed 'Difference Topology'. Grainge, I., Buck, D. and Jayaram, M. (2000). Geometry of site alignment during Int family recombination: antiparallel synapsis by the Flp recombinase. J. Mol. Biol. 298: 749-764.
7. 2000-2002. The orientation of strand exchange within the recombination complexes formed by Flp and Cre is determined by the asymmetry in the protein and not by sequence asymmetry in DNA. Dr. Ian Grainge, with help from Ms. Shailja Pathania, employs artificially symmetrized DNA substrates to show that they are still treated as asymmetric within the synapse, leading to strand exchange only in the antiparallel mode. Grainge, I., Pathania, S., Vologodskii, A., Harshey, R. M., and Jayaram, M. (2002). Symmetric DNA sites are functionally asymmetric within Flp and Cre site-specific DNA recombination synapses. J. Mol. Biol. 320: 515-527.
8. 2001-2003. Five negative supercoils are sequestered by a three-site interaction in the phage Mu transposition complex. Ms. Pathania, in collaborative work with Dr. Rasika Harshey's group, uses 'difference topology' to figure out the number of DNA crossings formed within the Mu transpososome. Of the total five crossings, two are between the right end of Mu and the enhancer DNA, two between the left and right ends and one between the left end and the enhancer. The same analysis is used to characterize the complexes at the stages of strand cleavage as well as the joining of target DNA. Pathania, S., Jayaram, M., and Harshey, R. M. (2002). Path of DNA within the Mu transpososome. Transposase interactions bridging two Mu ends and the enhancer trap five DNA supercoils, Cell 109: 425-436. Pathania, S., Jayaram, M. and Harshey, R. M. (2003). A unique right end-enhancer complex precedes synapsis of Mu ends: the enhancer is sequestered throughout transposition. EMBO J. 22: 3725-3736.
Plasmid Partitioning
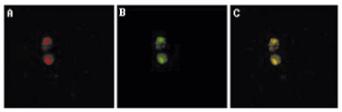
1. 1997-2000. The cell biological tools for studying the segregation of the yeast 2 micron circle are established. The notion that plasmid and chromosome segregation are coupled is postulated. Dr. Velmurugan, assisted by Mr. Yong-Tae Ahn, Ms. Xian-Mei Yang and Ms. Shwetal Mehta, demonstrates that the dynamics and kinetics of the 2 micron plasmid segregation parallel those of chromosome segregation. In conditional chromosome segregation mutants, the plasmid tends to segregate in tandem with the bulk of the chromosomes. Velmurugan, S., Ahn, Y. T., Yang, X-M., Wu, X-L., and Jayaram, M. 1998. The 2 micron plasmid stability system: Analyses of interactions among plasmid- and host-encoded components. Mol. Cell. Biol. 18: 7466-7477 Velmurugan, S., Yang, X. M., Chan, C. S., Dobson, M. and jayaram, M. (2000). Partitioning of the 2-micron circle plasmid of Saccharomyces cerevisiae. Functional coordination with chromosome segregation and plasmid-encoded Rep protein distribution. J. Cell. Biol. 149: 553-566.
2. 2000-2002. The plasmid has evolved mechanisms to steal components of the chromosome segregation pathway. Ms. Shwetal Mehta and Dr. Velmurugan discover that the plasmid recruits the yeast cohesin complex, required for the pairing of sister chromatids and their subsequent separation and movement to daughter cells, specifically to its partitioning locus. Overall, the results suggest cohesin mediated pairing of replicated plasmid clusters as a critical step in segregation. Mehta, S., Yang, X. M., Chan, C. S. M., Dobson, M., Jayaram, M., and Velmurugan, S. (2002). Coupling between 2 micron plasmid and chromosome partitioning in yeast: the plasmid stability system purloins the yeast cohesin complex. J. Cell Biol. 158: 625-637. (see also News section of J Cell Biol. August, 2002).
|