Prateek Gupta and David Hall
Botany 331 Spring 1997
A newly characterized plant DNA binding domain, called WRKY after its conserved sequence, contains a novel C2 -H2 zinc finger. The studied proteins with this domain are transcription factors for unrelated genes. We isolated and partially sequenced an A. thaliana cDNA clone and found the DNA sequence encodes a putative DNA binding protein with a C2 -H2 zinc finger characteristic of the WRKY domain. . After Northern Blot analysis, we determined that we sequenced 0.7 kilobases of a 1.2 kilobase gene. The partial characterization was sufficient to suggest a role for the protein to serve as a DNA binding protein and possibly as a transcription factor. Early data suggests that cellular damage increases levels of the mRNA encoded for by the gene. A method for the characterization of the protein's function and the gene's promoter sequence is discussed.
Key Words: transcription factor, C2 -H2 zinc finger, WRKY domain
The structural genes in eukaryotes, those genes that encode protein sequences, are transcribed by RNA polymerase II. Although it is an extremely large, multisubunit complex, RNA polymerase II often needs many specific proteins, called transcription factors, to selectively transcribe a gene specific to the temporal and spatial needs of the cell and/or tissue. These transcription factors are also necessary to surpass the basal rate to create the specific gene product in a sufficient quantity. (10) Transcription factors have various attributes, which allows them to have the necessary diversity to ensure separate control of the thousands of genes in eukaryotic genomes. These DNA binding proteins can bind promoters-DNA sequences which are immediately upstream of the transcribed region. In addition, transcription factors can bind enhancers--sequences which can be upstream, downstream or within introns of the transcribed region (10). Scientists group transcription factors according to their DNA binding motif(s); the three common motifs being the zinc finger, leucine zipper and helix-turn-helix.
In 1994, Sumie Ishiguro and Kenzo Nakamura characterized a sweet potato transcription factor called SPF1 that binds upstream of three different genes. Because SPF1 had no homologies to any known proteins, Ishiguro and Nakamura suggested that this protein might have a new type of DNA binding domain (8). Further study by Rushton, Macdonald, Huttly, Lazarus and Hooley revealed a consensus between SPF1 and other proteins with a similar binding domain, consisting of a conserved 24 amino acid sequence WRKYGQKxxKxxxxPRxYxx and a potential zinc finger C-X4-5 -C-X22-23-H-X-H. (15) This sequence was named the WRKY domain, and the sequence(s) to which they bind are known as W boxes. (16). The WRKY family is thought to be a broad group of plant DNA binding proteins, and plants with representatives of this family include parsley, wild oat, sweet potato, rice, turnip, cucumber, and A. thaliana . These proteins are involved in unrelated processes which include hormonal regulation, sucrose regulated gene expression and gene expression in defense mechanisms (16).
The second part of the WRKY domain is the novel C2 -H2 zinc finger. A zinc finger is a DNA binding motif that includes histidine and cysteine residues that are stabilized by zinc ions (11) Although the zinc finger in the WRKY domain has this feature, it also has unusual spacing between the histidine and cysteine residues, making it different than formerly characterized zinc fingers (15) Various experiments show that using zinc chelating agents such as 1,10-o-phenanthroline or EDTA destroy the binding ability of this domain, whereas EGTA, which chelates calcium, has no effect. (11).
Partially sequencing an unknown Arabidopsis thalania gene sequence has yielded a sequence that, once translated, contains a highly repeated zinc finger motif. The homology between this unknown zinc-finger motif and other known protein sequences is restricted to a stretch of 24 highly conserved amino acids followed by the WRKY zinc finger motif. Although the unknown sequence shares the zinc finger motif with other transcription factors, this does not suggest similarity in function of the transcription factors. Roles for these transcription factors can range from regulation of pathogenesis-mediated pathways to metabolism. Outside of the DNA-binding motif, the protein sequences vary, which provides an explanation for the different roles.
Isolation of unknown cDNA in pZL1, Subcloning and Minipreparation
The cDNA clone was obtained from the Arabidopsis Biological Resource Center and it included an unidentfied cDNA insert. The plasmid was isolated from DH5a E.Coli by a modified alkaline lysis method (1,13). The pZL1 plasmid was digested with EcoRI and HindIII (Promega) to isolate the insert of interest (14). Next, the insert was subcloned into a pBluescript vector (Stratagene). The pBluescript vector containing the unknown insert was transformed into XL1 Blue E.Coli , and then selected transformants were screened by PCR, using T3 and T7 primers and then by miniprep (7)(20)(21).
DNA sequencing and analysis
The Sanger method (21) and the Sequenase Version 2.0 DNA Sequencing Kit (24) were used to sequence the denatured dsDNA pBluescript plasmid. Primers T3 and T7 were used, and the sequencing was run on a Long Ranger Gel (12). Automated sequencing was performed on the pZL1 insert using T7 and SP6 primers. The structural analysis and sequence searches were performed using BLAST, FASTA, Oxford Molecular Group, and Hastings Software packages. The homologous protein sequences--accession numbers L44134 (SE71), U58520 (WRKY2), X92976 (ZAP1), Z48429 (ABF1), and G30038 (SPF1)--were taken from GenBank, and the sequence comparison was performed using ClustalW.
Nucleic acid isolation
Genomic DNA was isolated by grinding A. thaliana tissue in liquid N2 and extracting using 2% SDS and phenol. (2) (26). A restriction digestion using EcoR1, HindIII, and BamH1 endonucleases (Promega) was performed, and the digested DNA was separated on a 0.7% agarose gel (19). Total RNA was isolated from A. thaliana using phenol/chloroform/isoamyl alcohol and TLE-SDS (3) (22). Samples were taken from stems, leaves, flowers, and whole plant, and wounding samples were taken from whole plant tissue 0, 1, 5, and 8 hours after wounding. The RNA was run on a formaldehyde 1.2% agarose gel.
Southern and Northern hybridizations
RNA and DNA were transferred onto Zeta-Probe nylon blotting membranes, and hybridizations were performed using our a-32P dATP radiolabeled cDNA insert as a probe (3) (4) (25). Washes were performed at high stringency (1% SDS, 40 mM Na2HPO4 pH 7.4, 1mM EDTA at 60° C) and at low stringency (5% SDS, 40 mM Na2HPO4 pH 7.4, 1mM EDTA at 60° C) (27) (18).
Insert analysis
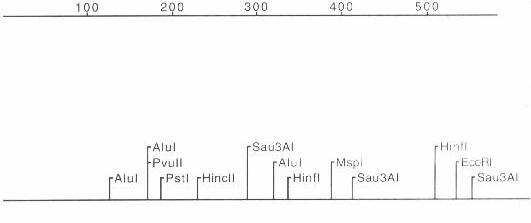
The insert's deduced restriction map is shown in Figure 1. The insert's end sequences were essentially nonoverlapping, implying the insert is larger than the sum of the two end sequences from the T7 and SP6 primers (approx. 1.2 Kb). The insert matched a published cDNA sequence, GenBank accession number T45479, from an A. thaliana expressed sequence library. The open reading frame that showed sequence homologies to previously characterized proteins is given in Figure 2.
Analysis of protein translation
The sequence from the T7 primer had the homology to the WRKY proteins, whereas the SP6 sequence gave no homologies to known proteins. The proteins containing this domain that showed the highest homology to our sequence were SE71 (9), WRKY2 (16), ZAP1 (11), ABF1 (15), and SPF1 (8), and these showed 75%, 78%, 75%, 80%, and 71% homologies, respectively. The zinc finger of the WRKY domain is shown in Figure 3.
Northern data
We found hybridization to a 1.2 kb expressed mRNA in our Northern blot. Among the three tissue types (flowers, leaves, and stems), the blot only showed a signal to flower tissue. We also found a signal to the wounded whole plant tissue which increased in intensity over time.
Southern data
The DNA gel blot revealed hybridization to a 0.6 kb EcoR1 fragment and to a 4.2kb BamH1 fragment. The low stringency wash (Figure 4) shows three related gene sequences (50-95% homology) that disappeared in the high stringency wash (not shown).

Figure 1 Restriction map and endonuclease cut site list for the 5' end of the cDNA insert. The primer T7 is to the right of the map. The sequenced part of the insert is the beginning 587 bp.

Figure 2 The deduced amino acid translation is shown below the nucleotide sequence. This is the open reading frame that showed homology to many plant DNA-binding proteins. The segment which contains the putative zinc finger characteristic of the WRKY domain is boxed, and the characteristic cysteine and histidine residues are marked.
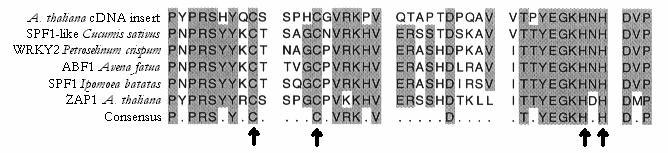
Figure 3 A multiple alignment of the homologous proteins showed a conserved sequence of 43 bp which includes the novel C2 -H2 zinc finger. The cysteine and histidine residues are marked.
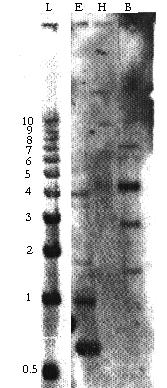
Figure 4 The Southern blot of the low stringency wash shows hybridization to a 0.6 kb EcoR1 fragment and a 4.2 kb BamH1 fragment. Approximately three additional genes sequences gave a weak signal to the probe.
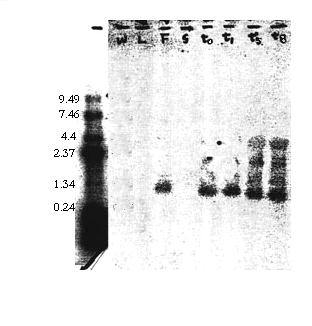
Figure 5 The Northern blot lanes are, from left to right: ladder (unmarked), whole tissue (W), leaves (L), flowers (F), stems (S), wounded 0hr. (t0), wounded 1hr.(t 1), wounded 5hr. (t5), and wounded 8hr. (t8 ). A signal to a 1.2kb transcript appeared in flower tissue and in response to wounding, and the wounding signal increased over time.
Sequencing the unknown cDNA sequence and identifying the best reading frame from open reading frame analysis yielded a nucleotide sequence of 327 bases, and a corresponding partial protein which has 109 amino acids (figure 2) Since no start codon was found, this protein is likely incomplete. Although the protein encoded by the unknown cDNA clone of interest contained a zinc finger binding domain found in other transcription factors like SE71, WRKY1 and ZAP1 (figure 3) , it did not share high homology outside of the zinc finger binding motif. The binding domain shows a higher homology in the domain to the other proteins than predicted by the C2-H2 model, indicating a conserved gene and possibly a similar mode of action.
Immediately flanking the zinc finger binding domain, there is also some homology between the known proteins and the protein encoded by the unknown cDNA. Comparison of the amino acid sequence between the different species shows high homology and further evidence of a highly conserved gene. These proteins represent a part of a new class of transcription factors, and since the initial blast searches did not retrieve a sequence with high homology, the protein encoded by the unknown cDNA could be as yet unidentified. This round of sequencing provided a partial sequence of the protein which would require further sequencing to obtain the primary amino acid sequence.
Northern blot analysis revealed expression of an mRNA that is nearly 1.2kb in size. In addition, it seems that this mRNA is localized in the flower, and increased expression is due to cellular damage. This evidence suggests a role for the encoded protein. Southern blot data indicates that the protein may have more than one copy in the genome, but a repeat of the experiment needs to be conducted to verify this. This data could shed light to a possible role of the unknown cDNA in Arabidopsis thaliana.
The WRKY binding domain and its novel zinc finger DNA-binding domain have not been well characterized. To characterize the encoded protein, a full length sequence of its expressed mRNA could be found using this insert as a probe.. The protein could then be expressed in a yeast vector, and its binding characteristics and promoter region could be studied. This could be done through the creation of multiple mutations in both the promoter sequence and the sequence of the protein. Also, the biological function of the gene could be studied by creation of a null mutation or through ectopic expression (11).
We thank K. Sathasivan for assistance in protocols and sequence analysis and Yew Lee for valuable help in the laboratory.
1. Birnboim, H.C. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods. Enzymol. 1983. 100:243-255.
2. Current Protocols in Molecular Biology. 2.21-2.33.
3. Ibid. 4.31-4.34.
4. DecaprimeII instruction manual. 1994 Ambion, Inc. Austin, TX.
5. Feinberg, A.P. and B. Vogelstein. 1983, 1984 Anal. Biochem. 183:6 and 137:266.
6. Gibco BRL. Transformation procedure for frozen competent cells.
7. Innis, M.A., D.H. Gelfand, J.J. Sninsky, and T.J. White. PCR Protocols. A guide to methods and applications. Academic Press. 1990. 3-13.
8. Ishiguro, S., K. Nakamura. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5' upstream regions of genes coding for sporamin and b-amylase from sweet potato. Mol. Gen. Genet. 1994. 244: 563-571.
9. Kim, D., S.M. Smith, and C.J. Leaver. A cDNA encoding a putative SPF1-type DNA-binding protein from cucumber. Gene. 1997. 185: 265-269.
10. Mathews, C.K., K.E. Van Holde. Biochemistry. The Benjamin/Cummings Publishing Company, Inc., 1995. 1061-1065.
11. Pater, S., V. Greco, K. Pham, J. Memelink, and J. Kijne. Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Research. 1996. vol. 24 no. 23. 4624-4631.
12. Protocol for Long Ranger Gel. By AT Biochem., Malvern, PA. 1-4.
13. Protocol modified by Drs. Paul Kreig and Doug Melton, Harvard University. Protocols and Applications Guide. 2nd Edition, Promega.
14. Qiagen Protocols and Applications Guide. Qiagen Inc., Chatsworth, CA. 1995.
15. Rushton, P.J., H. Macdonald, A.K. Huttly, C.M. Lazarus, and R. Hooley. Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of a-Amy2 genes. Plant Mol. Bio. 1995. 29:691-702.
16. Rushton, P.J., J.T. Torres, M. Parniske, P. Wernert, K. Hahlbrock, and I.E. Somssich. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO Journal 1996. vol. 15. no. 20. 5690-5700.
17. Sambrook, J., E.F. Fritsch, and T. Maniatis. Molecular Cloning-A Laboratory Manual. Cold Spring Harbour Press. 1989. 6.30-6.31.
18. Ibid. 10.13-10.17.
19. Ibid. 9.31-9.51.
20. Ibid. 14.1-14.21
21. Sanger, F., S. Nicklen, and A.R. Coulson. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 1977. 74:5463-5467.
22. Schuler, M.A. and R.E. Zelinski. Methods in plant molecular biology. Academic Press, Inc., New York. 1989. 1-9.
23. Ibid. 89-96.
24. Sequenase Version 2.0 DNA sequencing kit. Step by Step Protocols. United States Biochemical. Cleveland, OH. 1-25.
25. Southern, E.M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. Journal of Molecular Biology. 1975. 98: 503-517.
26. Tai, T., and S. Tangsley. Plant Mol Biol. Reporter. 1991. 8:297-303.
27. Zeta-Probe Blotting Membranes. Instruction Manual. BioRad. Hercules, CA.
We can be contacted through the Botany Department, Dr. Sathasivan, or directly.