Identification of a cDNA encoding an annexin-like protein in Arabidopsis thaliana.
Ryan S. Baldwin, Nicholas M. Vuong, Carter D. Shackelford, and K Sata Sathasivan1
1To whom correspondence should be written at the following address:
Painter 2.38, Botany Dept. University of Texas Austin, TX 78712
Abstract
Annexins are a superfamily of calcium-binding trans-membrane proteins found in protozoans and eukaryotes, but have yet to be completely described functionally. This large class of proteins have been implicated in deterring peroxidase activity (Gidrol et al.1996) and in adhesion properties of cell membranes (Morgan and Fernandez 1995). Annexin proteins in plant and animal species have been classified into more than twenty subfamilies. Through DNA sequencing methods and homology search, we have discovered a gene that codes for a protein similar to annexins in the plant, Arabidopsis thaliana. Southern hybridization was performed to detect the size and abundance of copy number of the gene in the A. thaliana genome. In addition, RNA was isolated from different tissues of the plant and used in a Northern hybridization to determine the quantity of message being transcribed and the size of the transcription. Results indicate that this annexin-like protein is a small gene family that is predominately expressed in the flower of the plant.
Introduction
The primary function of annexins is not known, however many subfamilies have been sequenced in many organisms including alfalfa (Pirck et al. 1994), maize (Battey at al. 1996), human (Tait et al. 1992), and chicken (Gerke and Koch 1990). Annexins have also been shown to play roles in binding extracellular calcium (Hong et al. 1981), cell fusion (Hong et al. 1981), cell proliferation (Munz et al. 1997), cell differentiation, and as adhesion molecules for various types of breast tumors (Ahn et al. 1997). This experiment was designed to isolate, replicate, sequence, and analyze an unknown fragment of A. thaliana DNA. When compared to a gene database, the DNA matched closely to many subfamilies of annexin. Experiments such as Southern and Northern hybridization were used to provide proof of the gene's existence in the genome of A. thaliana and to shed some light on how and where this gene is expressed.
Materials and Methods
Arabidopsis cDNA was obtained in the plasmid pZL1 from a cDNA library. After growing up in a Luria-Bertaini (LB) broth for 16 hours, bacteria cells containing the pZL1 plasmid and Arabidopsis insert were pelleted and used in standard alkaline-lysis DNA isolation protocol (Current Protocols). The plasmid was cut with EcoRI, BamHI, HindIII, and a combination of EcoRI and HindIII restriction enzymes. The restriction produced a 1.5 kb fragment using EcoRI and HindIII. This insert was cleaned using a gel extraction kit (Quiagen) and cloned into the pBluescript KS+ and transformed into DH5a competent bacterial cells (USB). After 2-3 hours growth, the cells were harvested for transformation during the lag phase. Once transformation was complete, these cells were plated on LB agar with ampicillin (100 mg/ml), IPTG (100nM), and X-Gal (2%) and grown overnight. White colonies were chosen and the cloned plasmids were isolated using a miniprep kit (Quiagen) and digested using EcoRI. Purified plasmid (using T3 and T7 primers), a white colony, and Red 6 DNA (positive control) were amplified using PCR. The PCR product was analyzed using agarose gel electrophoresis and sequenced using the sequencing facility at the University of Texas at Austin, which uses an ABI fluorescence sequencer (Perkin-Elmer). A 700 bp sequence with a poly A tail was sequenced and analyzed using the BLAST server on the NCBI Internet web page. Southern and Northern hybridizations were performed on DNA and total RNA, respectively. A. thaliana genomic was used for DNA and RNA was isolated from different tissues of the plant (whole, leaf, stem, and flower) and from a wounding time course. The wounding time course tissue consisted of whole plants injured at 0, 1, 5, and 8 hours. Probes were generated by purifying PCR product from primers and random nucleotides and using a random primer labeling kit (Ambion) and a-32P dATP (50mCi/reaction). The probe was used for both hybridizations. Genomic DNA was run out on a 1.2% agarose gel and blotted onto a nylon membrane and hybridized (Current Protocols). The RNA was treated in the same manner except the agarose gel contained formaldehyde as a denaturant.
Results
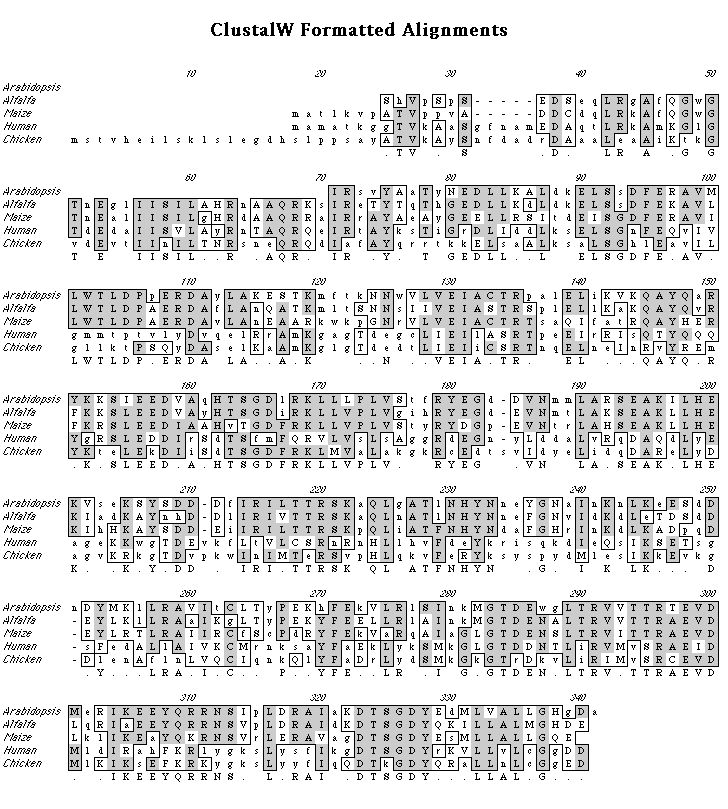
Once ligation of the insert into the pBluescript was complete, the transformation efficiency of the vector into the DH5a competent cells was 106 CFU/mg. A restriction map analysis showed cut sites for HincII and HindIII (selected restriction enzymes were used). Open reading frame analysis on the sequence gave a 269 protein. BLASTp results on the protein gave many strong matches (high score=668, P-value=10-133) to the various subfamilies of annexin. MACVECTOR was used to generate a ClustallW protein alignment comparing the A. thaliana sequence with annexins from alfalfa (Pirck et al. 1994), maize (Battey at al. 1996), human (Tait et al. 1992), and chicken (Gerke and Koch 1990). Hydrophobicity and motif analyses were performed to show the possibility of the protein having trans-membrane and calcium-binding attributes. The hybridization probe for the Southern and Northern analyses gave 2.77x108 CPM after synthesis and 9.76x107 CPM after a spin purification column giving 35% incorporation. The concentration for the probe was 2.44x106 CPM/ml hybridization solution. Four bands were present in the Southern blot suggesting that this sequence is part of a small gene family. The Northern hybridization showed three 1.34 kb bands in the one, five, and eight hour wounding time course tissues with the most abundant signal in the five hour tissue. One 1.34 band was present in the flower tissue, but the stem, leaf, and whole tissues were blank.
Discussion
Annexins are found throughout phyla including certain protozoans (Morgan and Fernandez 1995). The majority of matches to the protein sequence was to several annexin subfamilies suggesting that this protein may have similar functions to that of annexins. Hydrophobicity tests were performed to see if this annxein-like protein showed the trans-membrane properties associated with annexins. The results of these tests show that the protein does have hydrophobic and hydrophilic domains. Further tests to show homology to annexins would be to perform an in vitro translation of the sequence and determine if the protein has the ability to bind extracellular calcium. Data from the Southern blot shows that there are few copies of this annexin-like sequence in the genome of A. thaliana. The data from the Northern hybridization suggests that this annexin-like protein is involved in some type of wounding response, however, it is difficult to ascertain the exact function of the protein without further tests. Ribonuclease protection assays have been performed on Annexin II subunits with Transforming growth factor beta1 (TGF-beta1)to show the expression of the annexin gene in proliferating cells at the edge of a wound (Munz et al. 1997) and similar tests could be performed with A. thaliana.. Many tests need to be performed on this protein to find out how it is used in Arabidopsis.
Acknowledgments:
The authors thank Ryo Kawazoe for preparation of the experiments.
REFERENCES
Pirck, M., Hirt, H., and Hebererle-Bors, E. (1994). The cDNA sequence encoding an annexin from Medicago sativa. Plant Physiol. 104 (4), 1463-1464
Gerke V. and Koch W. (1990). The cDNA sequence of chicken annexin II. Nucleic Acids Res. 18, 4246
Tait, J. F., Smith, C., Frankenberry, D. A., Miao, C. H., Adler, D. A., and Disteche, C. M. (1992). Chromosomal mapping of the human annexin IV (ANX4) gene. Genomics 12, 313-318
Battey, N. H., James, N. C., and Greenland, A. J. (1996). cDNA isolation and gene expression of the maize annexins p33 and p35. Plant Physiol. 112 (3), 1391-1396
Current Protocols in Molecular Biology
Gidrol, X., Sabelli, P. A., Fern, Y. S., and Kush, A. K. (1996). Annexin-like protein from Arabidopsis thaliana rescues delta oxyR mutant of Escherichia coli from H2O2 stress. Proc. Natl. Acad. Sci. U.S.A. 93 (20), 11268-11273.
Munz, B., Gerke, V., Gillitzer, R., and Werner, S. (1997). Differential expression of the calpactin I subunits of annexin II and p11 in cultered keratrinocytes and during wound repair. J Invest Dermatol 108 (3): 307-312.
Hong K., Duzgunes, N., and Papahadjopoulos, D. (1981). Role of synexin in membrane fusion. Enhancement of calcium-dependent fusion of phospholipid vesicles. J Biol Chem 256 (8): 3641-3644.
Ahn, S. H., Sawada, H., Ro, J. Y., and Nicolson, G. L. (1997). Differential expression of annexin I in human mammary ductal epithelial cells in normal and benign and malignant breast tissues. Clin Exp Metastasis 15 (2):151-156.
Morgan, R. O. and Fernandez, M. P. (1995). Molecular phylogeny of annexins and identification of a primitive homologue in Giardia lamblia. Mol Biol Evol 12 (6): 967-979.
TGATCCGCAG CGTTTATGCA GCTACCTACA ATGAGGATCT TCTCAAAGCA TTAGACAAAG AGCTTTCTAG CGACTTTGAG AGAGCTGTGA TGTTGTGGAC TCTTGATCCA CCAGAGAGAG ATGCTTATTT GGCTAAAGAA TCCACCAAGA TGTTCACCAA GAACAATTGG GTTCTTGTTG AAATCGCTTG CACAAGGCCT GCTCTTGAGC TTATCAAGGT CAAGCAAGCT TACCAAGCTC GATACAAGAA ATCAATCGAG GAAGATGTCG CGCAACACAC ATCTGGTGAC CTTCGTAAGC TCTTGCTTCC TCTTGTGAGC ACTTTCAGGT ATGAAGGAGA TGATGTGAAC ATGATGCTTG CAAGATCTGA AGCTAAGATA CTTCACGAGA AGGTCTCAGA GAAATCTTAC AGTGACGATG ACTTCATCAG AATCTTGACA ACAAGAAGCA AAGCACAGCT CGGTGCAACA CTCAACCACT ACAACAACGA GTATGGAAAC GCCATTAACA AGAACTTGAA GGAAGAGTCG GACGACAATG ACTACATGAA ACTACTAAGA GCTGTAATCA CATGTTTGAC ATACCCTGAG AAGCATTTTG AGAAGGTTCT TCGTCTATCA ATCAACAAAA TGGGAACAGA CGAATGGGGA CTAACCCGAG TCGTGACTAC ACGAACTGAA GTTGACATGG AACGCATCAA AGAGGAATAT CAGCGAAGAA ACAGCATTCC TTTGGACCGT GCTATCGCCA AAGACACTTC TGGTGACTAT GAGGACATGC TTGTTGCTCT TCTCGGACAT GGCGATGCTT GAAACTGTTT CAACTTTCGA GTTCCTCCTT TCTCTTACTG CATGGTTTGT TTTAAATAAA AGAGTTGTGA AACTGGTTCT GCAACTATTT ATCAATGATC GTTTGAGTTT GTTAAAAAAA AAAAAAAA
Protein Sequence (beginning with base #3):
IRSVYAATYNEDLLKALDKELSSDFERAVMLWTLDPPERDAYLAKESTKMFTKNNWVLVEIACTRPALELIKVKQAYQARYKKSIEEDVAQHTSGDLRKLLLPLVSTFRYEGDDVNMMLA RSEAKILHEKVSEKSYSDDDFIRILTTRSKAQLGATLNHYNNEYGNAINKNLKEESDDNDYNKLLRAVITCLTYPEKHFEKVLRLSINKMGTDEWGLTRVVTTRTEVDMERIKEEYQRRN SIPLDRAIAKDTSGDYEDMLVALLGHGDA
Picture Gallery