Review
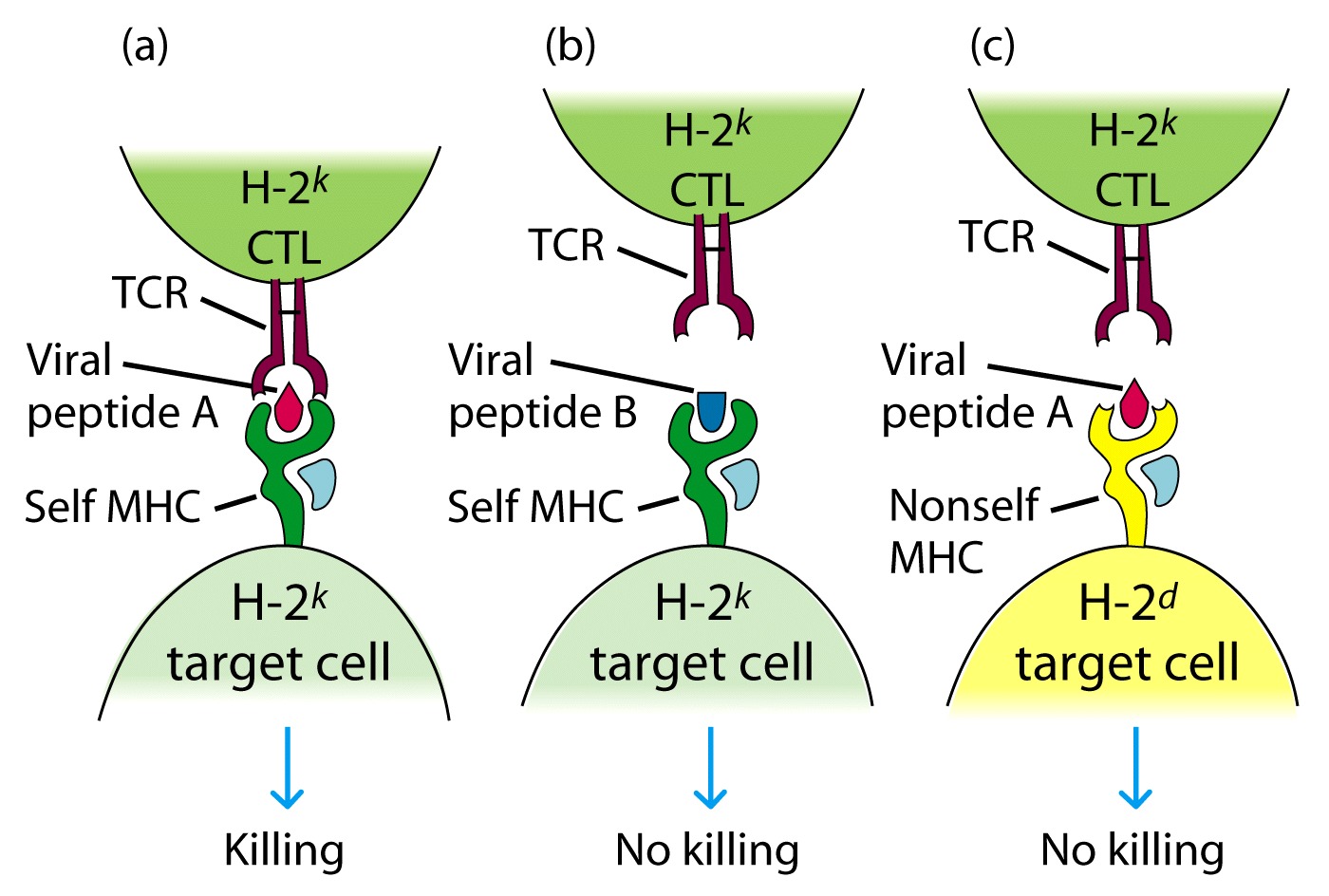
Zikernagel
MHC self restriction

Jim Allison (MD Anderson, Smithville, TX)
T CELL RECEPTOR
The structure of the T cell receptor was not elucidated until the 1980s. It was much more difficult to isolate the TcR than Ig because the T cell does not secrete its receptor and because the receptor is specific for both antigen and MHC.
Monoclonal antibodies and nucleic acid probes were both vital to the purification / isolation of the TcR.
The TcR was isolated using monoclonal antibodies against T cell clones. In other words, T cells derived from a single T cell bearing a single type of TcR. Just as in the isolation of Ig, a homogeneous preparation of TcRs was required.
Radiolabeled membrane of T cell and directed Ab against the membrane (were trying to find a distinct marker for T cells)
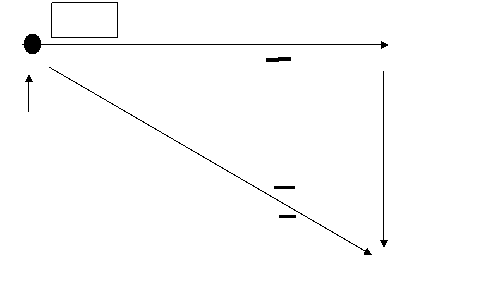

2 bands that were seen under reducing conditions suggested a heterodimer
Later determined to be α, β
The Monoclonal Ab recognized one protein on the TCR and didn't react with B cells
Q. Why was this important?
Later on Allison generated Ab that reacted with only some T cells but not all.
It was concluded that he had previously made Ab to the C region on the TCR and later the V region on the TCR
Based on these findings it was proposed that
TCR = 2 chains α, β that were made up of VDJ/C and VJ/C regions
STRUCTURE OF T CELL RECEPTOR
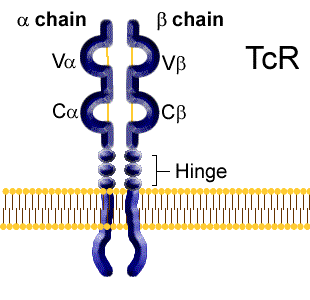
The T cell receptor is a heterodimer composed either of alpha and beta or gamma and delta polypeptide chains. Amino acid sequencing analysis shows a surprising similarity to the domain structure of the Igs. Each chain has a variable region domain and a constant region domain (designated Va and Ca, Vb and Cb etc.).
Three complementarity determining regions which appear to be equivalent to the CDRs in Ig heavy and light chains have been identified in both the alpha beta and gamma delta chains.
The variable region domains
of alpha & beta or gamma & delta come together to form the antigen
binding cleft.
The TcR has been visualized by X-ray crystallography and is quite similar in the
way in which the antigen binding site is formed by the CDRs of Ig variable
region domains.
The TcR heterodimer has a mw
of ~ 85,000 - 90,000 D.
The 2 polypeptide subunits are ~40,000 D each
IDENTIFYING AND CLONING THE TCR GENES
Marc Davis
Hedrick and Davis
Used the Approach of Subtractive Hybridization
Assumptions:
1) TcR genes expressed in T cells but not B cells (mRNA for the TcR will not be
found in B cells)
2) mRNA for the TcR would be found on polyribosomes (attached to RER)
3) T cell Receptor genes will be rearranged in mature T cells but not in other
cell types
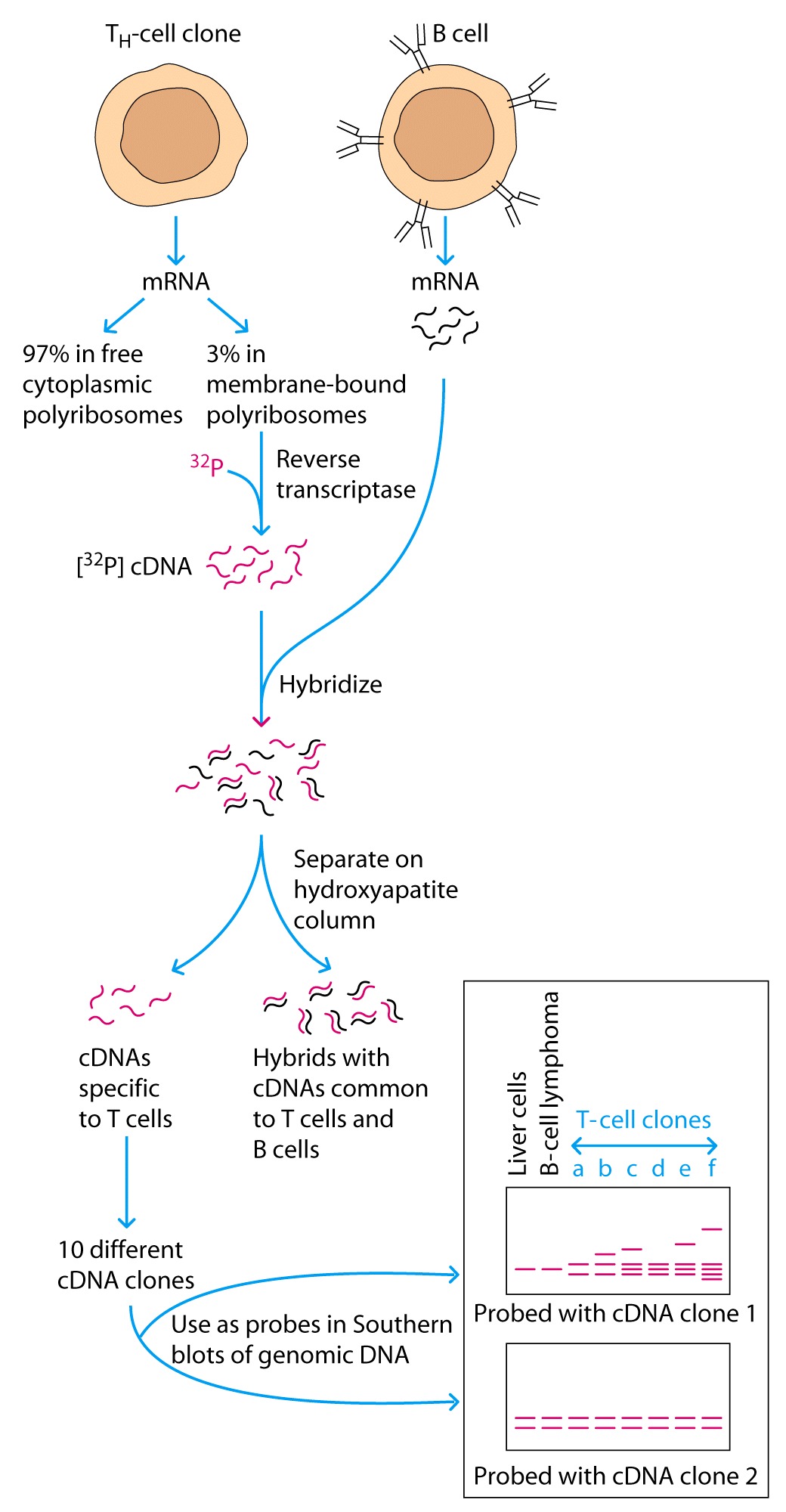
The investigators isolated the polyribosomal fraction from a
Th cell clone and used reverseb transcriptase to make 32P
labeled cDNA probes.
As
shown in Figure
1. Isolating
radioactively-labeled cDNAs (made using reverse transcriptase) that correspond
to mRNAs expressed in T cells but not in B cells;
2. Cloning
those T cell-specific cDNAs into plasmids;
3. Doing
Southern hybridization using each cloned cDNA as probe to test whether the DNA
segments recognized by the probe is in the same sized fragment in embryonic or
liver DNA as it is in T cells. They assumed that as is the case for antibody
genes, segments encoding TCR genes must be rearranged in T cells in order for a
T cell to commit to producing one TCR. If the probe detects different
hybridizing fragment(s) in T cells than in liver DNA, gene rearrangement must
have occurred and the cDNA is a candidate for encoding a TCR chain.
Summary
Mark
Davis and Stephen Hedrick at the NIH used a technique called subtractive
hybridization to try to isolate cDNAs --- pieces of DNA complementary to
mRNAs --- that encoded TCR chains. Their experiment resulted in isolation of a
cDNA encoding what came to be known as the TCR b
chain.
TCR genes
Germ-line configuration
Four loci – a,
b, g, and d. Expression of TCR genes involves rearrangement,
similar to the Ig genes.
The a and d loci are on
the same chromosome; the b
locus is on a different chromosome; the g
locus is on a different chromosome.
In Mice
There are 100 Va, 50 Ja,
1Ca.
The rearrangement of the a locus eliminates the d locus.
There are 10 Vd, 2Dd, 2Jd,
1Cd.
There are 20-30 Vb, followed by two repeats of 1Db, 6Jb, and 1Cb.
There are 7Vg, followed by three Jg-Cg
repeats.
Rearrangement – V(D)J
recombination.
gene rearrangement for a and b.
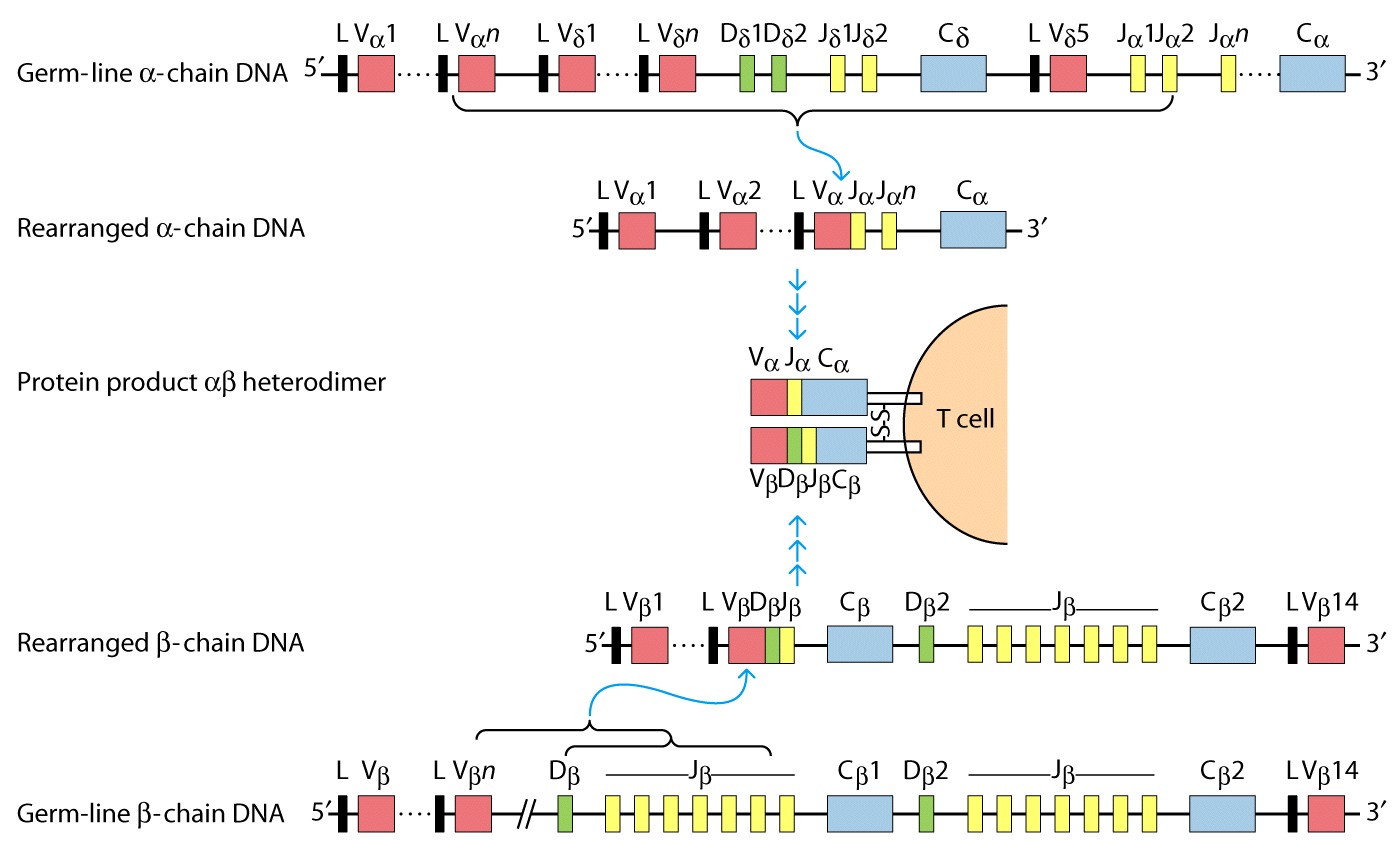
Same mechanism as in B cells – (similar heptamer and
nonamer sequences), Rag 1/2.
Generation of TCR
diversity
· Mechanisms: combinatorial joining; alternative joining for b and d; junctional flexibility; P region nucleotide addition; N region nucleotide addition (0-6 nucleotides in a, b, g, and d).
·
In contrast to Ig
genes, there is no somatic mutations in the TCR genes.
· The TCR has to recognize a large number of processed antigens, and a relatively small number of self-MHC.
TCR/CD3 complex
CD3 is required for the
surface expression of the TCR. Also, the CD3 is required for the initiation of
the transduction signals.
CD3 is formed by 5 invariant chains that form 3 types of dimers: ge, de, zz,
g, d,
and e – have Ig-like extracellular domains, TM domain, and 40 aa
intracellular domains.
z
and h – have only 9 aa extracellular, TM domain, long cytoplasmic domains
(113-155 aa).
The TM
domains contain negatively charged aa which interact with the positively charged
TM of the TCR.
The
intracellular domains contain ITAM motifs (g, d,
and e
have one, z
and h
have 3).
Accessory molecules
CD4
and CD8:
CD4 is
a 55 kD with 4 extracellular Ig-like domains, a transmembrane domain, and a long
cytoplasmic tail containing serine (which can be phosphorylated);
·
CD8
is a heterodimer ab; the MW of the chains is 30-38 kD, the chains contain
a single extracellular domain, a transmembrane domain, and a cytoplasmic tail
with 25-27 aa containing several phosphorylation sites.
The extracellular
domains of CD4 bind to the b2
of class II MHC;
·
The extracellular
domains of CD8 bind to the a3
of class I MHC.
·
The binding to
MHC increases the avidity of the interaction between the T cell and the
classI/classII-bearing cell approx. 100 times.
·
Both CD4 and CD8
associate through the intracytoplasmic tail with Lck (a protein tyrosine kinase
functional in T cells).
·
CD2, LFA-1,
CD28, CD45R
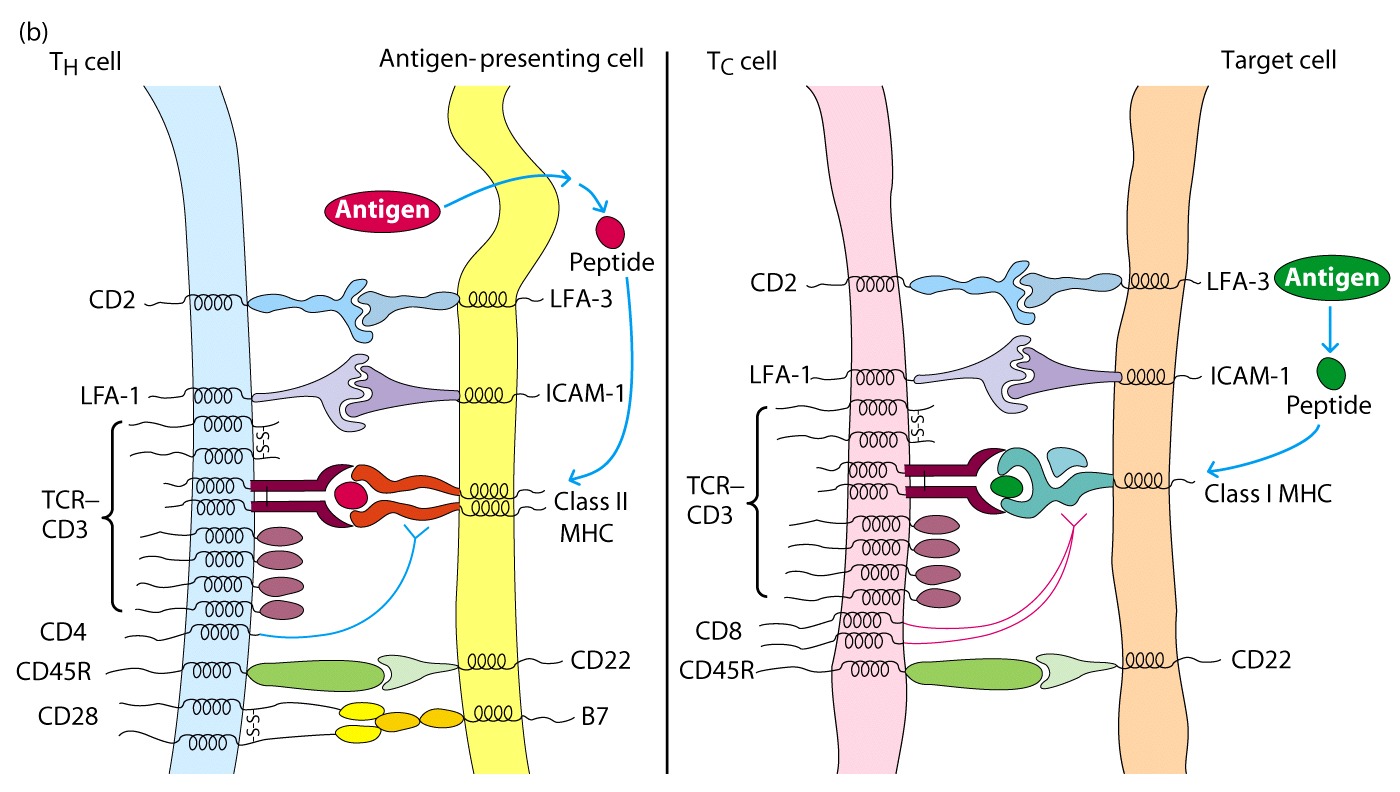
These molecules do not
interact with the MHC, but bind to other ligands on APC and strengthen
adhesion
CD28 on CD4+ T cells
interacts with B7 on APC and this interaction serves as a costimulatory signal
for T cell activation.
Without B7 there will be no response generated
Superantigens are compounds which can activate all T cells with specific sequences in their receptors. They will activate many more clones than will be activated with a specific antigen, but fewer than will be activated by a lectin.
Superantigens work by binding to both the TCR from a T cell and
the MHC molecule from an antigen-presenting cell - however, they bind to
completely different sites than those used by normal antigens. Thus, they
can bind a TCR with any antigen specificity to any MHC molecule.
Superantigens include the substance responsible for toxic shock syndrome and some staphlococcal enterotoxins. By binding to and activating such large numbers of T cells, they cause the release of relatively vast amounts of cytokines, sometimes with disasterous results.
Superantigens are viral or bacterial proteins that bind simultaneously to the Vβ domain of the T cell receptor and to the α chain of the MHC class II
Crosslinkage of a TCR and a class II MHC molecule by a superantigen produces an activating signal that induces T-cell activation and proliferation
