
|
Project 4: Exploiting Site-specific Recombination to Solve Basic and Applied Problems in Biology A. Altered specificity Flp variants
(Chen et al. 2002, Mol. Cell)
Site-specific recombination is, in principle, a potent tool for directed DNA rearrangements, including gene therapy. While target specificity is a great advantage of the system, it also imposes strong limitations on where it can be applied. Creating novel recombinases with altered specificities can overcome this impediment. We have employed tools of mutagenesis and directed evolution to create variants of the Flp recombinase that act on new DNA targets. Enlarging the substrate range for site-specific recombination has strong implications in biotechnology.
B. DNA topology: Analyzing DNA paths in complex systems
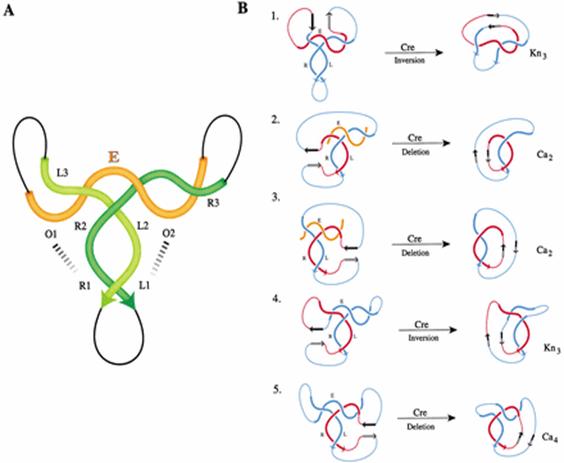
We use topological methods to analyze the consequence of DNA rearrangements mediated by recombinases and related enzymes. Our present understanding of the geometry and topology of the Flp reaction (and related systems) has formed the basis of an analytical tool called 'difference topology'). The method allows one to trace the path of two DNA distant segments when they are brought together within elaborate protein-DNA complexes (such as those involved in replication, transcription, DNA repair and DNA transposition). The rationale of difference topology is to set up the uncharacterized synapse, and then seal off the DNA crossings within it by tying up the DNA with the help of site-specific recombination. We can then use physical methods to count these crossings preserved in the DNA knots and DNA links generated by the recombination. The triumph of 'difference topology' is illustrated by the characterization of the phage Mu transposition complex (in collaboration with the Harshey laboratory; see Figures above). |
|
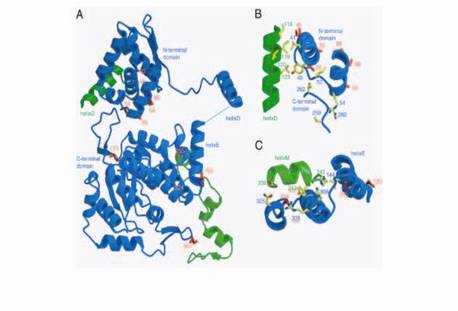
Locations of Mutations in Altered Specificity Flp Variants |