|
Project 3: Site-specific Recombination: Biochemistry and Mechanisms
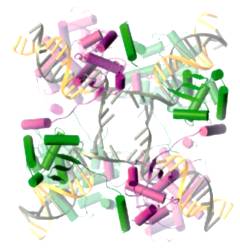
(Chen et al. 2002, Mol. Cell; figure kindly supplied by Dr P. A. Rice, Univ. Chicago)
We have solved nearly all of the biochemical and mechanistic details of the Flp recombination reaction: (1) how do the individual monomers within the tetramer interact with each other and with the DNA; (2) how do they coordinate the strand cutting and joining steps of recombination; and (3) why does the reaction follow a two step mechanism, first exchanging one pair of strands and then resolving the Holliday junction intermediate. Recent structural studies from other laboratories have confirmed our findings. We wish to use the recombinase active site as a model for understanding how complex active sites evolve from considerably simpler ones. We have revealed two cryptic ribonuclease active sites within Flp using appropriately designed synthetic substrates. The mechanistic details of the RNA cleavage activities are being studied. It is likely that the active site of the present day Flp arose from an elementary nuclease active site via a topoisomerase active site. Whereas the 2 micron plasmid is packaged as minichromatin within the yeast nucleus, all of the in vitro studies on recombination have been carried out on naked DNA molecules. We plan to carry out a careful analysis of the recombination reaction in vivo as well as study the reaction in vitro using DNA substrates that have been packaged into nucleosomes. |
|
Flp Recombination Complex |